Concepts
Architecture
Habitat in its simplest form is a two tier application: an agent process running as a daemon on each machine being examined and a client process to look at the data.
Long term or high sample rate data can be moved off to a specialist repository, called harvest, which also acts a reporting portal and an analytics engine. Harvest is covered in a separate set of documentation available at https://www.systemgarden.com/harvest and provides another optional tier to the architecture.
The following section describes the architecture of common deployments and the major concepts needed to understand habitat.
Single Host: ghabitat + clockwork
In its simplest form, habitat collects data and displays the timeseries on the same machine. Collection is carried out by a daemon program by the name of clockwork, which runs many periodic jobs, each of which collects data or carries out a calculation. The default configuration takes runs system collection jobs and inserts them into a central ringstore datastore.
A visualisation tool, such as ghabitat queries the daemon (or reads the datastore directly) and collects the data for tabular display or graphical plotting.
This scenario is found in one-off testing, when using single machines or running additional instances of habitat that are independent of a general collection infrastructure.
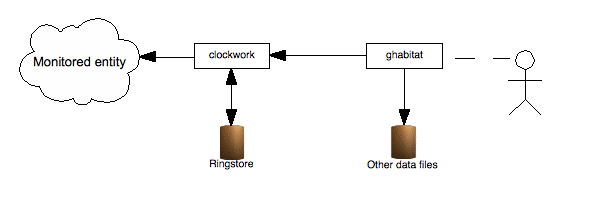
Illustration
1: Habitat running on a single machine, monitoring its own host.
Ghabitat displays information obtained from a data file or from the
clockwork collection agent.
Many Hosts: ghabitat + Many Clockworks
For small networks of machines with a handful of data viewers, habitat may be configured using peer level connections. By adding additional hosts to ghabitat's set of choices, a user may select to visualise data from remote machine.
To keep performance high, ghabitat only ever uses one connection at a time and polls at the same frequency of the sample interval. The connections are stateless, so the clockwork instances have no overhead in their role as servers of data. Additionally, the protocol is based on HTTP (but on a non standard port), that it allows for easy routing and checking in network infrastructures.
The result is a balance between containing demand and low administration.
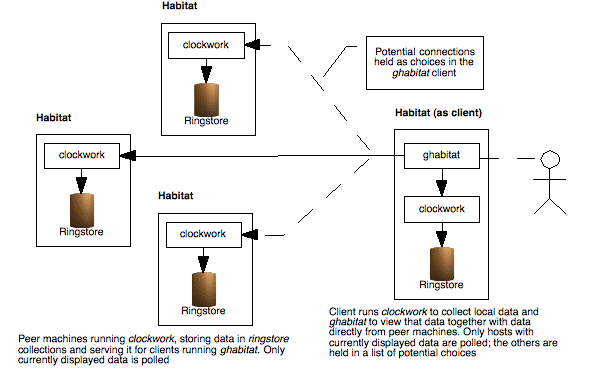
Illustration
2: Several instances of habitat, one of which is acting as a
visualisation tool. Whilst many connections can be in place,
ghabitat only polls for currently displayed data and at the same
rate it is sampled, thus reducing the overall load to a minimum.
Many Hosts with Repository: ghabitat + clockwork + harvest
For larger configurations or when there are many potential viewers, an additional component is employed to remove load from monitored systems and increase scalability. This is a central repository containing an archive of machine data and is implemented using the harvest product, also available from system garden (https://www.systemgarden.com/harvest).
Data may be sent directly to harvest using the route system, although there are many advantages in indirectly posting data. Habitat does this by default when it replicates many pieces of data at periodic intervals, thus batching the updates and minimising another potential bottleneck.
With a repository present, ghabitat will use that source to obtain data for all hosts and potentially many other centrally computed statistics. In this case, there is no additional load on the monitored servers.
Only unreplicated data would need to be collected from the monitored servers, generally the case where very recent data had been collected but the replication process had not been carried out. The process of replication is configurable, both time frequency and the data rings sent and received.
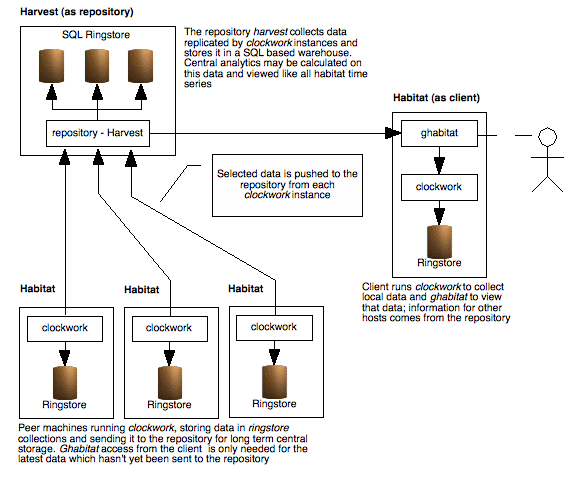
Illustration
3: Top level architecture of habitat and harvest, showing several
clients running habitat, one of which is viewing data with ghabitat,
and a single harvest repository. Using harvest lessens the load on
the the monitored machines, stores detailed long term data and
provides organisational wide analytics
The two can communicate by HTTP or by sharing the data file to which clockwork writes. Both these protocols are network friendly and allow multiple viewers to see the same data, or a single viewer to see many data sets.
Extensible Collection Methods
Within clockwork is the ability to extend collected data and the methods by which it can be collected. Data capture is scheduled using a single table (clockwork's job table) which may be different for each instance of clockwork. By adding or changing the jobs different data can be collected at custom time intervals. Each job runs a method, with several ones included in the standard distribution. New methods can be defined by adding code to clockwork at run time in the form of shared objects that have a standard interface. This is the most efficient way of adding new collection and computation capabilities, as the minimum number of processes are involved and overhead is kept to a minimum.
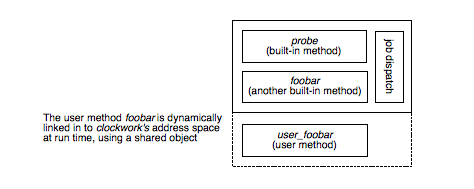
Illustration
4: The user method foobar is dynamically linked in to clockwork's
address space at run time, using a shared object
System vs. Private Clockwork Instances
On a single machine, many instances of clockwork can run, but only one can provide the dependable system service, which is able to serve data over the network using the standard port. Typically, a system-wide instance would cover most eventualities, but for some issues more detail profiling is required, such as heavily monitoring an application under test. In these cases, additional, private clockworks can be run, saving data to their own data files. These files can be viewed in real time or saved for later examination using the standard tools
Storage & Transport Integration
Habitat provides several ways of viewing collected data. Chief among them is habget, a command line tool to extract data for use in Unix filter pipelines, and ghabitat, a graphical application written in Gtk+. A more sophisticated product for larger installations and grids is under development by System Garden. It is cross platform and provides many advanced features (see separate documentation and the website https://www.systemgarden.com for more details).
Data Format
All data in habitat is tabular and can be expressed externally using a format called the Fat Headed Array, an example of which is shown below. It is tab delimited and similar in function to CSV (comma separated values) data, except that it can also contain sets of information attached to each column. These are known as info rows and are printed over several lines below the single column name row.
The single row of column names and the zero or more info rows, form an extended header block that is terminated by two or more dashes (--) on a single line. In the example above, the dashes have been extended to form a ruler line of the width of each column, similar to the convention of a SQL table display. Following the ruler is the tabulated array of values, which may be any sequence of characters excluding tab (\t) and the double quote (“). In summary, The fat headed array must have the same number of columns through out each row, but will be one more for the second and successive headers rows (the info rows).
Data Collection
Data is gathered by habitat's agent, clockwork, which can pull the data from a source or can have data pushed directly into the data store or other data stream.
In order to pull data, clockwork (habitat's agent) has a table of jobs that it executes at regular intervals (described later). By default these jobs run probes, that collect all manner of system information and send it to a data stream called a route. These routes usually address local persistent data storage, so that information sent to a route will end up being saved.

Illustration
5: The components involved in polling (pulling) monitored activities
or statistics and storing them inside habitat. Ringstores are
conventionally used as they are self-contained and require little
maintenance
Data can also be pushed on to routes independently of clockwork, by using an API or the command line tool, habput. This will take text, in the format of a Fat Headed Array, and will send it to the route address specified.
A time series of data is built up by repeatedly executing a probe (or receiving FHAs from external tools) to store a sequence of tables. Each table defines data over periods of time and is assigned a time stamp, sequence number and duration. This is often expressed in a tabular context by using the special columns _time, _seq and _dur.
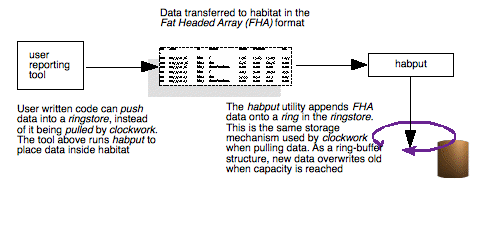
Illustration
6: The components involved in pushing data from user written
utilities or applications into habitat's storage system. Usually,
this will involve the creation of data in a Fat Headed Array (FHA)
format and its insertion into a local ringstore using habput
Each probe operation or FHA store will typically store data relating to a single point in time. However, when importing or reporting sets of data, the data series is concatenated into a single table and uses the_seq column to distinguish between samples.
A table of values are used for each sample, so that multiple instances may be expressed with out the use of excessive columns. For example, if habitat gathers information about storage, the FHA may look like the following.


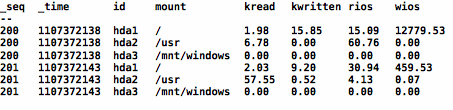


In the example above, there are two sets of three lines, with each set sharing the same sequence number: 200 and 201. These rows belong to the same sample, share the same time stamp but have different values for id, which is the instance key. In the case of storage, the instance key is a subset of the device name. Thus, to get a time series for a particular disk (say hda1), select the rows with id=hda1 and sort on _seq.
Data Addressing
Habitat and harvest share the same addressing scheme but have different implementations, suiting their particular design goals. Habitat generally uses a ringstore whereas Harvest uses a SQLringstore, which is based on more heavy weight database technology. However, as the route mechanism in habitat connects to both systems of storage, data is universally accessible across the product range.
To identify the different types of storage, a full route address contains a prefix, ending in a colon (:). For ringstores, the prefix is rs: and for the SQLringstore in the harvest repository the prefix is sqlrs:.The type (or driver) prefix is very similar to the URL format used in web browsers and have some similar capabilities. The list of drivers currently supported are as follows:-
-
rs:
Ringstore
sqlrs:
SQLringstore
file:
Plain file. When writing will append to existing file
fileov:
Plain file. When writing will destructively overwrite existing file
http:
https:Hypertext transfer protocol and secure hypertext transfer protocol, the encrypted variety. Most standard URL formats are supported. Requires configuration variables to be set up to allow communication though proxies and to use accounts.
stdin:
stdout:
stderr:Standard input, output and error. No further address required
none:
Output will be disposed of. No further address required
Data Storage
Data is generally collected in a local data store named a ringstore. This is a lightweight but structured storage system that has a lower impact on the system than a full SQL database. It is based in key-value block storage, currently implemented with GDBM.
Six dimensions are required to uniquely address a single datum, which are shown in the table below. Only the first two are needed to insert data, that of file and ring names: all the other details come from the inbound data.
-
File name
(or host name)The file name that holds the data. There is usually one file per machine with default central collection, although with personal collection, this becomes a user choice. With sqlrs (SQLringstore) from the harvest repository, this dimension is the host name.
Ring name
A collection of data tables sharing the same type or schema. Typical values are sys for system information, io for storage and net for network data. Note that the schema is not fixed or predefined: see below for details.
Duration
The interval between samples in seconds or 0 for an irregular sampling period. If omitted then the data is consolidated over all available durations (see below).
Column
Optional selection of a column name, which is able to return a single attribute of data (sets of columns may also be possible).
Sequence
Optional single or range of sequence numbers, uniquely identifying rows belonging to the same sample. Identified by pre-pending s= to the range.
Time
Optional single or range of time values, that can be used to extract a time series from a ring of data. Identified by pre pending t= to the range. All time values are in the Unix time_t format (seconds from 1/1/1970 or the epoch)
Ringstore & SQLRingstore
The native format for habitat is the ringstore, which is based on a GDBM key-value storage database and is typically held on storage local to the machine being monitored. Harvest is based on a SQL database and makes its data available using an HTTP format using a web servelet. The format is known as SQLringstore and has a very similar addressing format to the local ringstore.
The general format for ringstore and SQLringstore is as follows:-
[sql]rs:file_or_host,ring[,duration[,column]][,s=srange][,t=trange]
Where
|
[sql]rs: |
Either rs: or sqlrs: for ringstore or SQLringstore respectively |
|
file_or_host |
The file name of the ringstore, conventionally ending with .rs to informally indicate its type. With SQLringstore this will be the host name of the machine that generated the data |
|
ring |
Ring name, such as io, sys or net |
|
duration |
Optional duration of samples, measured in seconds. 0 (zero) is taken to mean an irregular event. If missing on reads, the consolidated view is taken; if missing on writes, the column _dur will be expected in the data |
|
column |
Optional column to extract when reading. Unused when writing |
|
s=srange |
Sequence range. srange is of the form from [ - [ to ] ] |
|
t=trange |
Time range. trange is of the form from [ - [ to ] ] |
The following are examples of addresses used by habitat:-
|
rs:myhost.rs,sys,60 |
Ringstore file called myhost.rs, returning the ring sys taken at 60 seconds interval |
|
rs:/var/lib/habitat/myhost.rs,sys,300 |
Ringstore file
called myhost.rs in the |
|
fileov:/home/fred/.habrc |
Flat text file, being the personal configuration of habitat for the user fred. The file is .habrc in fred's home directory. The type fileov: is chosen as the file is replaced each time it is generated by habitat. |
|
When used as an output, data is appended to the file mylog.txt. Used as inputs, fileov: and file: types are identical in method. |
|
|
sqlrs:myhost,sys,300,*,s=428- |
As an input, data is extracted from the repository (system myhost, ring sys at duration 300 seconds) and returned as a table. Only sequences 428 and greater are returned. |
Local Data Storage
In a standard configuration, habitat stores centrally collected data in a ringstore structure, which is held in a single file. The file is called hostname.rs and is held in var in the application directory (for the .tar distributions) or /var/lib/habitat (for the RPM distributions).
Individual users may also collect customised data for their own use, which will not be stored in the central ringstore file. Typically, they will use this data in addition to the central information by mounting both files within a visualisation tool such as ghabitat.
The central file is also used to provide peer data access and data replication (see below).
Peer Data Access
The normal method to access local and remote data is to query the agents directly on each monitored machine. The agent (clockwork) implements a network server to satisfy queries from the front end ghabitat and other tools. When given a query, it accesses the central habitat ringstore on the local machine in order to return the results. For security reasons, it is not possible to use this method to access any other file.
In ghabitat your local host's data will appear under 'local host' in the choice tree. To connect to other hosts and get their data, select File->Host, type the name of the machine and click 'direct'. If successful, an entry for that machine will appear in the choice tree, under the branch 'my hosts'.
It is also possible to export data files using a file sharing protocol such as NFS or CIFS and mount them on remote machines. Using this technique for centrally collected ringstores may impact the ability the speed and reliability of data access due to the nature of file locking and network bandwidth. However, this may be the most appropriate way of sharing custom data collected by individual users as the files may not be as busy.
In ghabitat, connect to ringstore files by selecting File->Open and navigating to the location of the data file. The file will appear in the choice tree under the branch 'my files'.
Remote Data Repository
Using the standard configuration, the central ringstore file will grow to around 50 MBytes; more if additional data is collected or retention periods are extended. Moreover, older data is summarised at a lower frequency than originally collected to save space.
To keep data for longer, a remote repository may be used to archive data and can be used as a data source within ghabitat just like ringstore files or host attachments. Such a repository is provided by the harvest product, with data stored in the SQLringstore format.
From ghabitat, the repository is used by mapping the host into your choice tree. There are two methods. Firstly, by directly referencing the host: selecting File->Host from the menu, typing the host name and keeping the repository button highlighted. The host will appear under the branch 'my hosts' in the choice tree.
The second method is by browsing the repository from the choice tree. Hosts in the harvest repository may be ordered by organisational structure. For example, a server in London's finance department may be reached using the following tree path:
repository->Finance->London->theserver
Regardless of the method, the tree structure below the hosts will show the data that has been transferred to the repository. See the Data Replication section for details of data transfer to the repository.
Data Replication
Habitat sends data o the harvest repository by a process called replication. It enables data that has been collected in the local ringstore to be synchronised with the repository and new data transferred.
However, replication may also work both ways by allowing centrally created data to be propagated down to satellite habitat instances. This is an ideal way of maintaining job tables or other data which needs to be independent of network connections.
Typically, replication runs once a day, but can easily be increased for sites with a policy of frequent archiving. The process is always initiated by clockwork using a job from its job table. If harvest or other similar tool wanted to pull data from habitat, clockwork's network service should be used using standard route conventions. Replication is not enabled in the standard configuration of habitat. Instructions to configure and enable it are contained in the Administration Manual.
Replication of data has a number of benefits, including:-
Data is backed up to an alternative location (the repository)
Visualisation or extraction tools can use the repository for the data source, saving capacity on the analysed machine
The repository can hold large quantities of data, which may be inappropriate to store locally on machines being analysed.
The repository can be a specialist database machine, with large storage and general purpose data handling
With the performance statistics of the whole enterprise in a single place, centralised analytics may be run in an efficient manner.
See System Garden's harvest repository for more details.
User Interfaces
A variety of visualisation and extraction tools exist for use with habitat, many of which are available in the package and are described below. In addition, System Garden provides other tools to enhance the interface which are available in separate packages. See https://www.systemgarden.com for more details.
Command Line
Habitat provides two command line utilities for data extraction and insertion. As such, they can be used in shell pipelines to build more complex commands.
habget extracts data from any supported route using and returns the data on stdout. See the manual later in this documentation for details. However, as an example:-
habget rs:/var/lib/habitat/myhost.rs,sys,60 | more
Will return the most recent data sample from the system probe and pipe the output into the utility 'more'. The data will appear as a table as shown in examples above. The file is the standard location under Linux for RPM installations.
habput is a method of inserting data onto a route for storage in a ringstore or SQLringstore. The manual for its use is later in the documentation, however an example is shown below:-
habput rs:myfile.rs,myring,0 <<END tom dick harry -- 1 2 3 4 5 6 END
Will read the table from stdin and send it to the ringstore myfile.rs, to be stored in the ring myring. The table will be scanned and checked to confirm that it is tabular before it is stored. It is timestamped with the current time and a sequence allocated depending on the existing contents of the ring.
The commands clockwork and killclock are used to stop and start the collection daemon on each machine. These are explained in a section below.
The remaining commands are covered by the Administration Manual:-
|
habedit |
Edits configuration tables within ringstores |
|
habmeth |
Runs clockwork methods from the command line |
|
habprobe |
Runs built-in data collection probes from the command line |
|
habrep |
Forces a replication cycle to take place |
|
irs |
Interactive ringstore utility, allowing administration of the data held in ringstore files |
Curses
The curses-based, dumb terminal tool is not supported in the current release.
Graphical
The main visualisation tool within the habitat suite is ghabitat. To start this, either select from the 'start' bar of your windowing environment or type ghabitat on the command line.
